Leave a Comment on Data Wrangling with R. อ่านการใช้งานเพิ่มเติมและดาวน์โหลด cheat sheet. Data wrangling with dplyr and tidyr cheat sheet tidy data foundation for wrangling in ma ma in tidy data set: each variable is saved in its own column syntax.
- Dplyr Cheat Sheet
- R Data Wrangling Cheat Sheet Excel
- R Data Wrangling Cheat Sheet Pdf
- R Cheat Sheet Pdf
10.1 Suffixes
| suffix | use when |
|---|---|
| _all | you want to apply the verb to all columns |
| _at | you want to apply the verb to specified columns |
| _if | you want to apply the verb to all the columns with some property |
10.2 Examples
10.2.1mutate(), summarize(), select(), and rename()
10.2.1.1 Named functions
| Verb | Example | Example explanation |
|---|---|---|
| summarize_all | summarize_all(mean) | finds the mean of all variables |
| summarize_at | summarize_at(vars(x, y), mean) | finds the mean of variables x and y |
| summarize_if | summarize_if(is.double, mean) | finds the mean of all double variables |
| mutate_all | mutate_all(as.character) | converts all variables to characters |
| mutate_at | mutate_at(vars(x, y), as.character) | converts variables x and y to characters |
| mutate_if | mutate_if(is.factor, as.character) | converts all factor variables to characters |
| rename_all | rename_all(str_to_lower) | changes all column names to lowercase |
| rename_at | rename_at(vars(X, Y), str_to_lower) | changes the names of columns X and Y to x and y |
| rename_if | rename_if(is.double, str_to_lower) | changes the names of double columns to lowercase |
| select_all | select_all(str_to_lower) | selects all columns and changs their names to lowercase (better to use rename_all()) |
| select_at | select_at(vars(X, Y), str_to_lower) | selects just columns X and Y and changes their names to x and y |
| select_if | select_if(is.double, str_to_lower) | selects just double columns and changes their names to lowercase |
10.2.1.2 Extra arguments
| verb | example | example_explanation |
|---|---|---|
| summarize_if | summarize_if(is.double, mean, na.rm = TRUE) | finds the mean, excluding NAs, of all double variables |
| summarize_all | summarize_all(mean, trim = 0.1, na.rm = TRUE) | finds the mean of all variables, exluding NAs. Removes the bottom and top 10% of values of each variable before computing mean |
10.2.1.3 Anonymous functions
| verb | example | example_explanation |
|---|---|---|
| summarize_all | summarize_all(~ sum(is.na(.))) | determines the number of NAs in each column |
| select_if | select_if(~ n_distinct(.) > 1) | selects only the columns with more than one distinct value |
10.2.2filter()
| verb | example | example_explanation |
|---|---|---|
| filter_all | filter_all(all_vars(!is.na(.)) | finds rows without any NAs |
| filter_all | filter_all(any_vars(!is.na(.)) | finds rows with at least one non-NA value |
| filter_at | filter_at(vars(x, y), all_vars(!is.na(.)) | finds rows where both x and y are non-NA |
| filter_at | filter_at(vars(x, y), any_vars(!is.na(.)) | finds rows where at least one of x and y is non-NA |
| filter_if | filter_if(is.double, all_vars(!Is.na(.)) | finds rows where all double variables are non-NA |
| filter_if | filter_if(is.double, any_vars(!Is.na(.)) | finds rows where at least one double variable is non-NA |
Approximate time: 75 minutes
The Tidyverse suite of integrated packages are designed to work together to make common data science operations more user friendly. The packages have functions for data wrangling, tidying, reading/writing, parsing, and visualizing, among others. There is a freely available book, R for Data Science, with detailed descriptions and practical examples of the tools available and how they work together. We will explore the basic syntax for working with these packages, as well as, specific functions for data wrangling with the ‘dplyr’ package, data tidying with the ‘tidyr’ package, and data visualization with the ‘ggplot2’ package.
All of these packages use the same style of code, which is snake_case formatting for all function names and arguments. The tidy style guide is available for perusal.
Adding files to your working directory
We have three files that we need to bring in for this lesson:
- A normalized counts file (gene expression counts normalized for library size)
- A metadata file corresponding to the samples in our normalized counts dataset
- The differential expression results output from our DE analysis using DESeq2
Download the files to the data folder by right-clicking the links below:
- Normalized counts file: right-click here
- Differential expression results: right-click here
Choose to Save Link As or Download Linked File As and navigate to your Visualizations-in-R/data folder. You should now see the files appear in the data folder in the RStudio file directory.
Reading in the data files
Let’s read in all of the files we have downloaded:
Tidyverse basics
As it is difficult to change how fundamental base R structures/functions work, the Tidyverse suite of packages create and use data structures, functions and operators to make working with data more intuitive. The two most basic changes are in the use of pipes and tibbles.
Pipes
Stringing together commands in R can be quite daunting. Also, trying to understand code that has many nested functions can be confusing.
To make R code more human readable, the Tidyverse tools use the pipe, %>%, which was acquired from the ‘magrittr’ package and comes installed automatically with Tidyverse. The pipe allows the output of a previous command to be used as input to another command instead of using nested functions.
NOTE: Shortcut to write the pipe is shift + command + M
An example of using the pipe to run multiple commands:
Dplyr Cheat Sheet
The pipe represents a much easier way of writing and deciphering R code, and we will be taking advantage of it for all future activities.
Exercises
Extract the
replicatecolumn from themetadatadata frame (use the$notation) and save the values to a vector namedrep_number.Use the pipe (
%>%) to perform two steps in a single line:- Turn
rep_numberinto a factor. - Use the
head()function to return the first six values of therep_numberfactor.
- Turn
Tibbles
A core component of the tidyverse is the tibble. Tibbles are a modern rework of the standard data.frame, with some internal improvements to make code more reliable. They are data frames, but do not follow all of the same rules. For example, tibbles can have column names that are not normally allowed, such as numbers/symbols.
Important: tidyverse is very opininated about row names. These packages insist that all column data (e.g. data.frame) be treated equally, and that special designation of a column as rownames should be deprecated. Tibble provides simple utility functions to handle rownames: rownames_to_column() and column_to_rownames(). More help for dealing with row names in tibbles can be found:
Tibbles can be created directly using the tibble() function or data frames can be converted into tibbles using as_tibble(name_of_df).
NOTE: The function as_tibble() will ignore row names, so if a column representing the row names is needed, then the function rownames_to_column(name_of_df) should be run prior to turning the data.frame into a tibble. Also, as_tibble() will not coerce character vectors to factors by default.
Exercises
Create a tibble called
df_tibbleusing thetibble()function to combine the vectorsspeciesandglengths.Change the
metadatadata frame to a tibble calledmeta_tibble. Use therownames_to_column()function to preserve the rownames combined with using%>%and theas_tibble()function.
Differences between tibbles and data.frames
The main differences between tibbles and data.frames relate to printing and subsetting.
Printing
A nice feature of a tibble is that when printing a variable to screen, it will show only the first 10 rows and the columns that fit to the screen by default. This is nice since you don’t have to specify head to take a quick look at your dataset. If it is desirable to view more of the dataset, the print() function can change the number of rows or columns displayed.
Subsetting
When subsetting base R data.frames the default behavior is to simplify the output to the simplest data structure. Therefore, if subsetting a single column from a data.frame, R will output a vector (unless drop=FALSE is specified). In contrast, subsetting a single column of a tibble will by default return another tibble, not a vector.
Due to this behavior, some older functions do not work with tibbles, so if you need to convert a tibble to a data.frame, the function as.data.frame(name_of_tibble) will easily convert it.
Also note that if you use piping to subset a data frame, then the notation is slightly different, requiring a placeholder . prior to the [ ] or $.
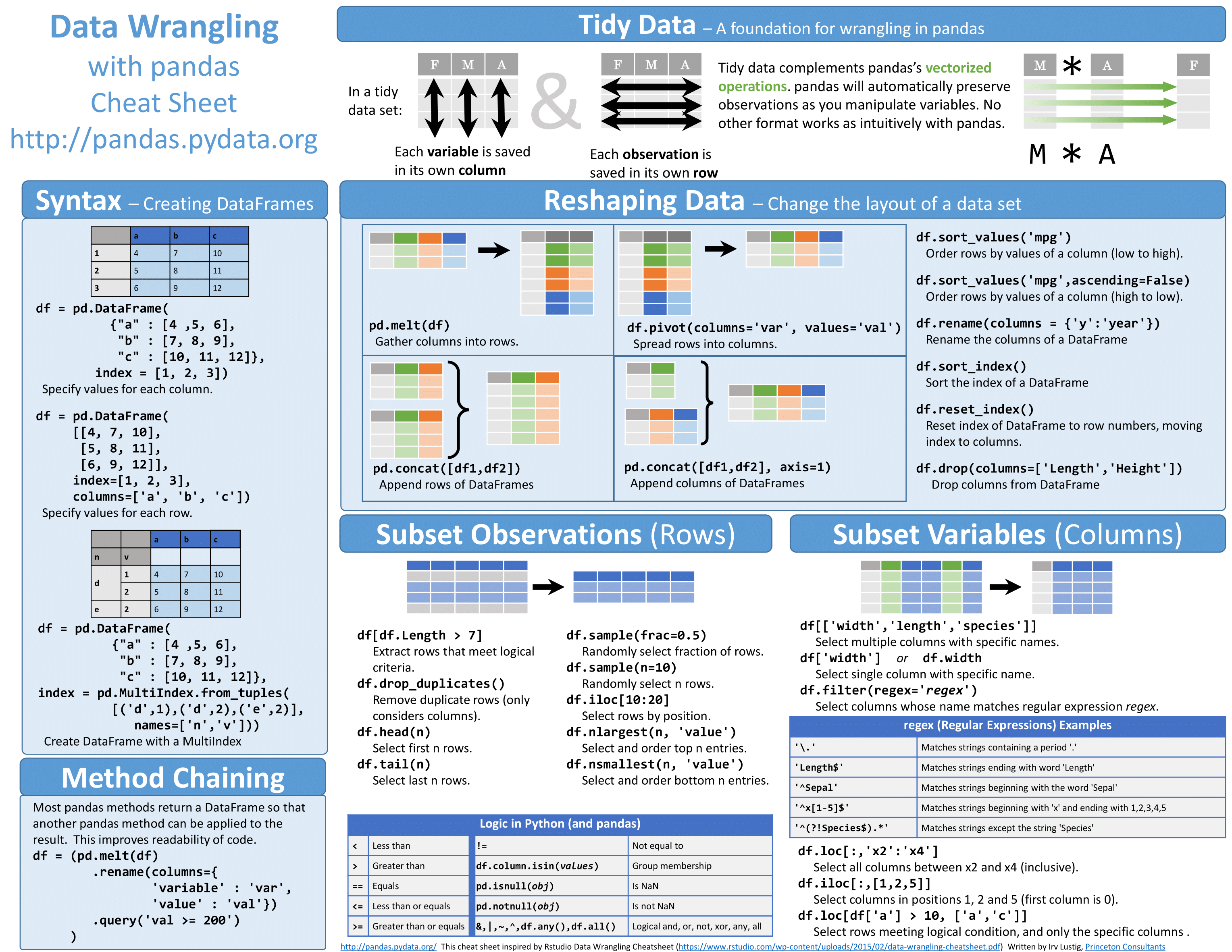
Tidyverse tools
While all of the tools in the Tidyverse suite are deserving of being explored in more depth, we are going to investigate only the tools we will be using most for data wrangling and tidying.
Dplyr
The most useful tool in the tidyverse is dplyr. It’s a swiss-army knife for data wrangling. dplyr has many handy functions that we recommend incorporating into your analysis:
select()extracts columns and returns a tibble.arrange()changes the ordering of the rows.filter()picks cases based on their values.mutate()adds new variables that are functions of existing variables.rename()easily changes the name of a column(s)summarise()reduces multiple values down to a single summary.pull()extracts a single column as a vector._join()group of functions that merge two data frames together, includes (inner_join(),left_join(),right_join(), andfull_join()).
Note:dplyr underwent a massive revision this year, switching versions from 0.5 to 0.7. If you consult other dplyr tutorials online, note that many materials developed prior to 2017 are no longer correct. In particular, this applies to writing functions with dplyr (see Notes section below).
select()
Jlg 1255 telehandler. To extract columns from a tibble we can use the select().
Conversely, you can remove columns you don’t want with negative selection.
arrange()
Note that the rows are sorted by the gene symbol. Let’s fix that and sort them by adjusted P value instead with arrange().
filter()
Let’s keep only genes that are expressed (baseMean above 0) with an adjusted P value below 0.01. You can perform multiple filter() operations together in a single command.
mutate()
mutate() enables you to create a new column from an existing column. Let’s generate log10 calculations of our baseMeans for each gene.
rename()
You can quickly rename an existing column with rename(). The syntax is new_name = old_name.
summarise()
You can perform column summarization operations with summarise().
Advanced:summarise() is particularly powerful in combination with the group_by() function, which allows you to group related rows together.
Note: summarize() also works if you prefer to use American English. This applies across the board to any tidy functions, including in ggplot2 (e.g. color in place of colour).
R Data Wrangling Cheat Sheet Excel
pull()
In the recent dplyr 0.7 update, pull() was added as a quick way to access column data as a vector. This is very handy in chain operations with the pipe operator.
_join()
R Data Wrangling Cheat Sheet Pdf
Dplyr has a powerful group of join operations, which join together a pair of data frames based on a variable or set of variables present in both data frames that uniquely identify all observations. These variables are called keys.
inner_join: Only the rows with keys present in both datasets will be joined together.left_join: Keeps all the rows from the first dataset, regardless of whether in second dataset, and joins the rows of the second that have keys in the first.right_join: Keeps all the rows from the second dataset, regardless of whether in first dataset, and joins the rows of the first that have keys in the second.full_join: Keeps all rows in both datasets. Rows without matching keys will have NA values for those variables from the other dataset.
To practice with the join functions, we can use a couple of built-in R datasets.
Tidyr
The purpose of Tidyr is to have well-organized or tidy data, which Tidyverse defines as having:
- Each variable in a column
- Each observation in a row
- Each value as a cell
R Cheat Sheet Pdf
There are two main functions in Tidyr, gather() and spread(). These functions allow for conversion between long data format and wide data format. The downstream use of the data will determine which format is required.
gather()
The gather() function changes a wide data format into a long data format. This function is particularly helpful when using ‘ggplot2’ to get all of the values to plot into a single column.
To use this function, you need to give the columns in the data frame you would like to gather together as a single column. Then, provide a name to give the column where all of the column names will be present using the key argument, and the name to give the column where all of the values will be present using the value argument.
spread()
The spread() function is the reverse of the gather() function. The categories of the key column will become separate columns, and the values in the value column split across the associated key columns.
Programming notes
Underneath the hood, tidyverse packages build upon the base R language using rlang, which is a complete rework of how functions handle variable names and evaluate arguments. This is achieved through the tidyeval framework, which interprates command operations using tidy evaluation. This is outside of the scope of the course, but explained in detail in the Programming with dplyr vignette, in case you’d like to understand how these new tools behave differently from base R.